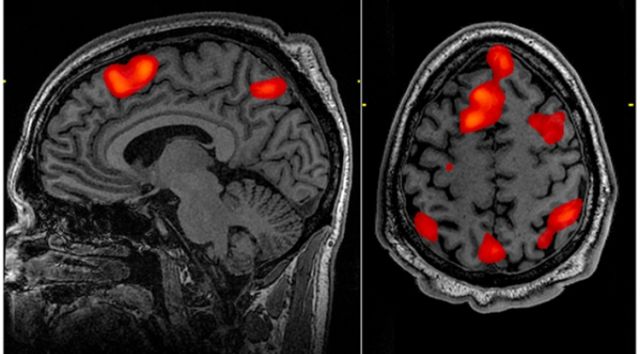
It's not an exaggeration to say that functional MRI has revolutionized the field of neuroscience. Neuroscientists use MRI machines to pick up changes in blood flow that occur when different areas of the brain become more or less active. This allows them to noninvasively figure out which areas of the brain get used when performing different tasks, from playing economic games to reading words.
But the approach and its users have had their share of critics, including some who worry about over-hyped claims about our ability to read minds. Others point out that improper analysis of fMRI data can produce misleading results, such as finding areas of brain activity in a dead salmon. While that was the result of poor statistical techniques, a new study in PNAS suggests that the problem runs significantly deeper, with some of the basic algorithms involved in fMRI analysis producing false positive "signals" with an alarming frequency.
The principle behind fMRI is pretty simple: neural activity takes energy, which then has to be replenished. This means increased blood flow to areas that have been recently active. That blood flow can be picked up using a high-resolution MRI machine, allowing researchers to identify structures in the brain that become active when certain tasks are performed.
But the actual implementation is rather complex. The imaging divides the brain up into small units of volume called voxels and registers the activity in each of these individually. Since these voxels are incredibly small, software has to go through and look for clustering—groups of adjacent voxels that behave similarly. The dead salmon results came because this software wasn't by default configured to handle the huge number of voxels imaged by current MRI machines. That meant even at 95 percent confidence, false positives were inevitable.
The new work, performed by a group of Swedish researchers, suggests that the software has other problems, too. The researchers took advantage of a recent trend toward making data open for anyone to use or analyze. They were able to download hundreds of fMRI scans used in other studies to perform their analysis.
Their focus was on scans of resting brains, typically used as a control in studies of specific activity. While these might show specific activities in some of the subjects (like moving a leg or thinking of dinner), there should be no consistent, systemic signal across a population of people being scanned.
The authors started with a large collection of what were essentially controls, randomly chose some to be controls again, and then randomly chose others to be an "experimental" population. They repeated this thousands of times, feeding the data into one of three software packages. The process was repeated with slightly different parameters in order to see how this affected the outcome.
The results were not good news for fMRI users. "In brief," the authors conclude, "we find that all three packages have conservative voxelwise inference and invalid clusterwise inference." In other words, while they're likely to be cautions when determining whether a given voxel is showing activity, the cluster identification algorithms frequently assign activity to a region when none is likely to be present. How frequently? Up to 70 percent of the time, depending on the algorithm and parameters used.
For good measure, a bug that has been sitting in the code for 15 years showed up during this testing. The fix for the bug reduced false positives by more than 10 percent. While good that it's fixed, it's a shame that all those studies have been published using the faulty version.
The authors also found that some regions of the brain were more likely to have problems with false positives possibly because of assumptions the algorithms make about the underlying brain morphology.
Is this really as bad as it sounds? The authors certainly think so. "This calls into question the validity of countless published fMRI studies based on parametric clusterwise inference." It's not clear how many of those there are, but they're likely to be a notable fraction of the total number of studies that use fMRI, which the authors estimate at 40,000.
The authors note that with current open data practices, it would be easy for anyone to go back and re-analyze the original work with the new caution in mind. But most of the data behind the already published literature isn't available, so there's really not much to be done here except to use added caution going forward.
PNAS, 2016. DOI: 10.1073/pnas.1602413113 (About DOIs).
reader comments
29