Fans of Douglas Adams’s Hitchhiker’s Guide to the Galaxy treasure the bit where a group of hyper-dimensional beings demand that a supercomputer tells them the secret to life, the universe and everything. The machine, which has been constructed specifically for this purpose, takes 7.5m years to compute the answer, which famously comes out as 42. The computer helpfully points out that the answer seems meaningless because the beings who instructed it never knew what the question was. And the name of the supercomputer? Why, Deep Thought, of course.
It’s years since I read Adams’s wonderful novel, but an article published in Nature last month brought it vividly to mind. The article was about the contemporary search for the secret to life and the role of a supercomputer in helping to answer it. The question is how to predict the three-dimensional structures of proteins from their amino-acid sequences. The computer is a machine called AlphaFold. And the company that created it? You guessed it – DeepMind.
Proteins are large biomolecules constructed from amino acid residues and are fundamental to all animal life. They are, says one expert, “the most spectacular machines ever created for moving atoms at the nanoscale and often do chemistry orders of magnitude more efficiently than anything that we’ve built”.
But these vital biomachines are also inscrutable because they assemble themselves into structures of astonishing complexity and beauty. (Illustrations of them make one think of what can go wrong when trying to wrap Christmas presents with those nice ribbons that only shop assistants can manage.) Understanding this “folding” process is one of the key challenges in biochemistry, partly because proteins are necessary for virtually every cell in a body and partly because it’s suspected that mis-folding may help to explain diseases such as diabetes, Alzheimer’s and Parkinson’s.
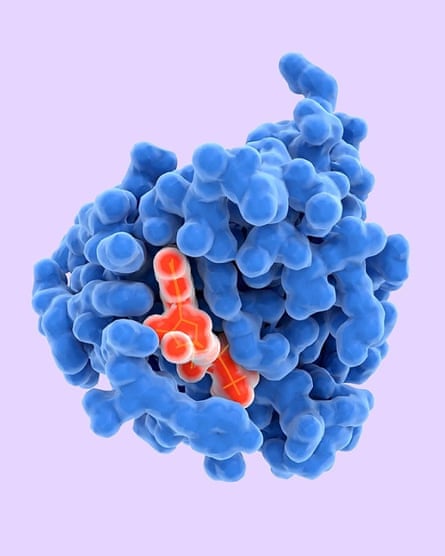
So the question “How do proteins fold?” is definitely worth asking. The traditional way of answering it was by lab-based x-ray crystallography, which is expensive and slow. So researchers have turned to building computer models that simulate the folding process and predict protein structures. For some years, specialists in the field have run a biennial competition in critical assessment of protein structure prediction (CASP), where teams are challenged to design computer programs that predict protein structures from amino sequences.
Two years ago, DeepMind, having conquered the board game Go, decided to take on the challenge, using the deep-learning technology it had developed for Go. The resulting machine was, predictably, named AlphaFold. At the CASP meeting last December, it unveiled the results. Its machine was, on average, more accurate than the other teams and by some criteria it was significantly ahead of the others. For protein sequences modelled from scratch – 43 of the 90 – AlphaFold made the most accurate prediction for 25 proteins. Its nearest rival only managed three.
These results seem to have had a seismic impact on many of the researchers present. The atmosphere and the implications were summed up in a remarkable blog post entitled “What Just Happened?” by Harvard’s Mohammed AlQuraishi, a world expert in the field. On the one hand, he was judiciously cautious about the contribution of the DeepMind team. It represented “substantial progress, more so than usual”. But does that mean the problem is solved or nearly so? “The answer right now,” he concludes, “is no. We are not there yet. However, if the [AlphaFold-adjusted] trend… were to continue, then perhaps in two CASPs, ie four years, we’ll actually get to a point where the problem can be called solved.”
On the other hand, AlQuraishi also discussed the existential angst generated by AlphaFold in the young scientists present at the event. Their underlying concern, he says, was “whether protein structure prediction as an academic field has a future, or whether… the best research will from here on out get done in industrial labs, with mere breadcrumbs left for academic groups”. Young biochemists will have to decide whether it’s good for their careers to continue working on structure prediction. For some (many?) of them, it may make sense to go into industrial labs, while for others it will mean staying in academia but shifting to entirely new problems that avoid head-on competition with DeepMind.
Underpinning all this, though, is a deeper question. Reaching a scientific explanation of how protein folding works will be a gigantic intellectual task. (In 1969, the molecular biologist Cyrus Levinthal formulated a famous paradox: as any protein can fold in an astronomically large number of ways it would take longer than the universe has existed for every configuration to be tested, yet most small proteins fold spontaneously in milliseconds. Nobody knows how.)
It’s conceivable that a machine-learning approach will soon enable us to make accurate predictions of how a protein will fold and this may be very useful to know. But it won’t be scientific knowledge. After all, AlphaFold knows nothing about biochemistry. We’re heading into uncharted territory.
What I’m reading
Sale of the century
Machine learning used to be an exotic technology. Now Timothy B Lee argues in Vox that it’s being commoditised. That may not be as good as it sounds.
What’s your poison?
If some algorithms can have harmful psychological effects on users, shouldn’t they be regulated like pharmacological drugs? There’s an interesting argument about it in Wired’s opinion section.
Progressive learning
Should “progress” be an academic subject? Read Diane Coyle’s thoughtful essay on the matter on the Project Syndicate website.

