Exploring and Cataloging the Diverse Human Microbiome
The human gut microbiome - the vast group of microbes that resides in our gut - plays an essential role in our health and well-being, studies have shown. But there is still a lot to learn about the relationship between humans and the many microbes that can be a part of the microbiome in our guts and other parts of our bodies. If we are to understand how specific microbes affect human health, it will be critical to identify and characterize every microbial species accurately. A new study reported in Cell involving an international team of scientists has aimed to catalog a greater number of the microbes that live in the human gastrointestinal tract.
“We genetically characterized and cataloged a large number of bacteria and archaea that are part of the human microbiome, but remained so far unexplored, uncharacterized, and undescribed,” said Dr. Nicola Segata, a human microbiome researcher. “We also observed that many of these microbes tend to be only rarely identified in Westernized populations, most probably as an indirect consequence of the complex industrialization processes.”
“It's the collection of bacteria, archaea, viruses, fungi, and parasites that populate human body sites such as the gut, the mouth, the skin, and the urogenital tract. The human microbiome is in symbiosis with our own cells and plays a key role for our health, for example, in the metabolism of dietary compounds, in regulating the so-called gut-brain-axis, in protecting us from pathogenic agents, and in modulating our immune system. It has also been recently shown that the microbiome is involved in the etiology of some cancers and in the success of anti-tumoral immunotherapy approaches,” explained Segata.
The team used a computational metagenomics approach in this study; they isolated all of the genetic information in a sample of saliva, skin, or stool, and sequenced the DNA to identify the microbes in the sample. The tool is outlined in the video above. The researchers used computational tools to reconstruct the genomes of the microbes from the data they obtained from their samples.
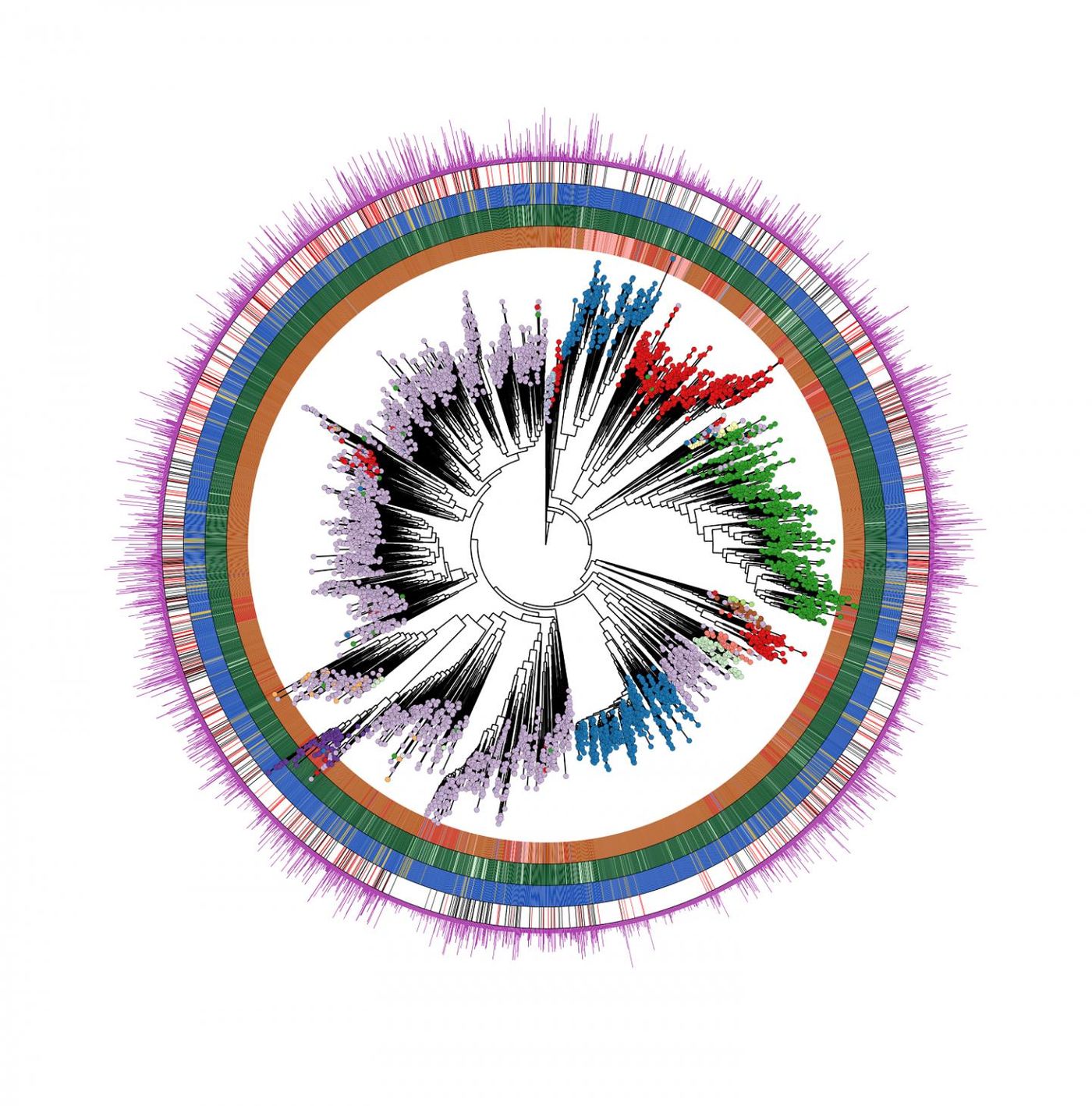
“Our findings are the result of a multidisciplinary team at CIBIO comprising microbiologists, statisticians, and computer scientists that identified a total of almost 5,000 microbial species recapitulating over 154,000 newly reconstructed genomes describing the human microbiome across ages, body sites, lifestyles, and diseases. Each of us is colonized by several hundreds of these species,” explained Segata. “But a large fraction of them (77 percent) were previously unknown. Many of these species are relatively rare, but some are very prevalent in human populations worldwide, and their discovery is the starting point for testing their potential role in autoimmune, gastrointestinal, and oncologic diseases. To obtain these results, we analyzed an extremely large dataset of publicly available and newly obtained microbiome samples spanning geography, population lifestyles, and age. Overall, we considered 9,428 human microbiome samples that have been studied using DNA sequencing technology called metagenomics.”
“The most common previously unknown candidate species, that we called ‘Cibiobacter qucibialis’ is the seventh most prevalent human-associated microbe in the worldwide population. We studied it by reconstructing more than 1,800 genomes. We think this species could be of particular relevance for further understanding the functions of the human microbiome,” said Segata.
“We specifically focused on non-Westernized populations that do not have access to [a] high-fat diet, common drugs including antibiotics, and do not live in highly sanitized environments. Many novel microbes discovered in non-Westernized populations across continents tend to be almost undetectable in Westernized populations. Our work thus enabled the study of such microbial species that could potentially be linked in the future with the increasing incidence of autoimmune diseases, allergies, and complex syndromes in the Westernized populations. It will thus be crucial to isolate, cultivate, and maintain these species that could be hypothetically reintroduced in Westernized populations with novel intervention strategies,” concluded Segata.
Sources: AAAS/Eurekalert! via University of Trento, Cell
-
APR 30, 2024Immuno-Oncology Virtual Event Series 2024
-
MAY 07, 20243rd International Biosecurity Virtual Symposium
-
JUN 06, 2024The Future of Scientific Conferencing
- See More